Version 1.4
PSICQUIC is meant to be a RESTful service. Hence, it is possible to retrieve data using simple URLs through HTTP.
SOAP can be too cumbersome for some users or developers as usually a client may be needed. With REST, none of this is necessary because it is possible to do a query through HTTP and get the data back. The simplest way to do it is to do queries using your browser, but REST is a convenient way to access the data for scripts.
Different methods are available using REST. It is possible to fetch data by using:
- One or many interactor/participant identifiers.
- One or many interaction identifiers.
- A MIQL flexible query.
The first two methods are just a convenient modification of the third method. The only difference is that the appropriate MIQL field is automatically added for you behind the scenes.
Other methods exist to get additional information about the service, such as specific properties or the version.
Differences from previous versions
From version 1.3
- Support all the fields of MIQL 2.8
- MITAB 2.8 is supported (combatibility with previous versions is maintained)
- RDF format support is no longer maintained
From version 1.2
- Support all the fields of MIQL 2.7
- XGMML format is available without any restrictions in number of interactions
- MITAB 2.6 and MITAB 2.7 are supported
- Now based on a SOLR index
- Bug fixed when exporting in Biopax
- RDF format support is no longer maintained
- Returns HTTP 400 error if we try to download more than 500 interactions in XML or Biopax in one request.
From version 1.1
- Data can be retrieved in a compressed form to save bandwidth.
- HTTP headers X-PSICQUIC-
*are not part of the response. - XGMML format available.
- Biopax format available
- RDF format support is no longer maintained
From version 1.0
- Methods have been added to retrieve properties about the service.
URL syntax
The structure of the URL to fetch data from PSICQUIC is this one:
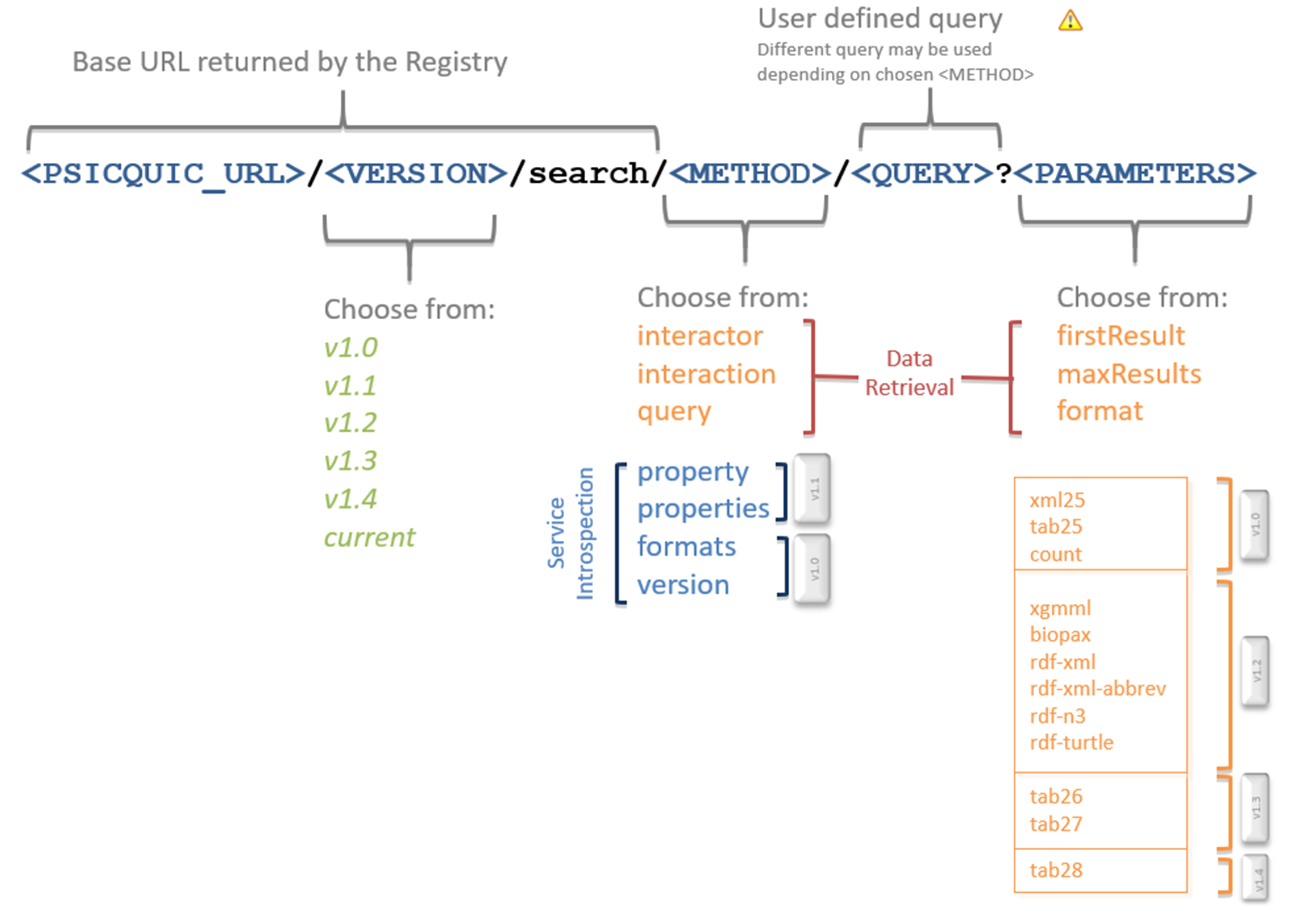
The URL contains different parts:
PSICQUIC_URL
The first element of the URL is, of course, the location of the web service. Check the page for the providers for specific addresses.
VERSION
At this moment there are 5 versions of PSICQUIC implemented. The possible values are:
| Version | Description |
|---|---|
| v1.0 | Version 1.0 of the REST PSICQUIC specification |
| v1.1 | Version 1.1 of the REST PSICQUIC specification |
| v1.2 | Version 1.2 of the REST PSICQUIC specification |
| v1.3 | Version 1.3 of the REST PSICQUIC specification |
| v1.4 | Version 1.4 of the REST PSICQUIC specification |
| current | Will map always to the most current version. It works as a symbolic link |
METHODS TO RETRIEVE MOLECULAR INTERACTION DATA
As we said before, there are different methods available to retrieve data:
| Method | Since spec version | Description |
|---|---|---|
| interactor | v1.0 | To search using interactor identifiers or aliases |
| interaction | v1.0 | To search using interaction identifiers |
| query | v1.0 | To search using MIQL queries |
METHODS WITH INFORMATION ABOUT THE SERVICE
These metadata methods return information about the service:
| Method | Since spec version | Description |
|---|---|---|
| property | v1.1 | To get the value of a property |
| properties | v1.1 | Lists all the available properties |
| formats | v1.0 | Lists the return formats supported by the service |
| version | v1.0 | Implementation version |
QUERY
This is the place where we define the actual query. Depending on the method different queries may be used. When searching for interactors or interactions, the query must be one or many identifiers separater by the operands AND or OR (e.g. facd AND brca2).
When using the query method, the query can be anything.
For users accessing RESTful PSICQUIC using scripts, the query must be properly encoded to avoid illegal URIs. For instance, all spaces in the query must be replaced by %20. Most programming languages contain already methods to do this. For clever web browsers, this is not necessary.
PARAMETERS
Whereas the rest of parts of the URL are mandatory, parameters are optional. We can use the parameters to choose the output format or to paginate the query. The parameters available in the reference implementation are the following:
| Parameter Name | Possible values |
|---|---|
| format | Format of the returned data. See the next section for the possible values |
| firstResult | Integer value that defines the first element to be shown. If the whole result set contains 100 interactions and we use firstResult=40, the first row to be returned will be the 40 |
| maxResults | Integer value that defines the maximum number of interactions to be returned. For instance, a value of maxResults=20 will limit the results to 20 interactions. |
It is possible to iterate through the results using "pages" by changing the value of firstResult while maintaining an immutable maxResults value. For example:
...?firstResult=0&maxResults=20
...?firstResult=20&maxResults=20
...?firstResult=40&maxResults=20
etc
If you don't want to show the whole results at once, this "pagination" method is very effective memory and speedwise.
Formats
This is the list of available formats:
| Format parameter | Since Spec | Description |
|---|---|---|
| tab25 | 1.0 | PSI-MITAB 2.5 |
| tab26 | 1.3 | PSI-MITAB 2.6 |
| tab27 | 1.3 | PSI-MITAB 2.7 |
| tab28 | 1.4 | PSI-MITAB 2.8 |
| xml25 | 1.0 | PSI-MIXML 2.5.4 |
| count | 1.0 | Just the total count |
| biopax | 1.2 | BioPAX - append -L2 or -L3 to the parameter if a specific level is desired |
| xgmml | 1.2 | XGMML format, used in Cytoscape) |
HTTP Headers
Since the spec 1.2, in order to help understanding the content of a response with results, some HTTP headers are used:
| Header name | Description |
|---|---|
| X-PSICQUIC-Count | Total count of results |
| X-PSICQUIC-Impl | Name of the implementation |
| X-PSICQUIC-Spec-Version | Version for the REST Spec used |
| X-PSICQUIC-Supports-Formats | List of supported formats |
For example:
curl -I "http://localhost:9090/psicquic/webservices/current/search/query/*?firstResult=0&maxResults=10"
HTTP/1.1 200 OK
Content-Length: 0
Content-Type: text/plain
Date: Wed, 18 Jul 2018 16:23:35 GMT
X-PSICQUIC-Count: 2
X-PSICQUIC-Impl: Reference Implementation
X-PSICQUIC-Impl-Version: 1.3.15-SNAPSHOT
X-PSICQUIC-Spec-Version: 1.4
X-PSICQUIC-Supports-Formats: xml25, tab25, tab26, tab27, tab28, biopax, xgmml, count, biopax-L2, biopax-L3
Server: Jetty(8.1.0.RC5)
Error status codes
REST relies on direct HTTP communication. Errors are reported using HTTP status codes in the header of the request:
| Status code | Description |
|---|---|
| 200 | Not an error. Request is OK |
| 406 | Format not supported |
| 400 | Too many results for exporting in XML and Biopax |
| 500 | Internal server error |
Some Examples
By Interactor
Getting the result for "brca2_human" in PSI-MITAB 2.7 format
Getting the result for "brca2_human" using the Uniprot accession P51587 in PSI-MITAB 2.7 format
The number of results in both cases is the same:
http://www.ebi.ac.uk/Tools/webservices/psicquic/intact/webservices/current/search/interactor/brca2_human?format=count http://www.ebi.ac.uk/Tools/webservices/psicquic/intact/webservices/current/search/interactor/P51587?format=count
By Interaction
- Getting the result for all the interactions which accession number contains "IM-16571-" in PSI-MITAB 2.7 format. These interactions are part of the publication with imex id IM-16571 (PubMed id "15710747")
By Query
Getting the result for "brca2" in PSI-MITAB 2.7 format
The same, in PSI-MI XML 2.5.4 format
It supports paging. For instance, search for "species:human" and get only the first 100 results
All of matrixdb in PSI-MITAB format
http://matrixdb.univ-lyon1.fr:8080/psicquic/webservices/current/search/query/*