This project hosts the PSICQUIC Reference Implementation, an implementation of the PSICQUIC Specification. A publication about PSICQUIC can be found in Nature Methods.
You can see PSICQUIC in action with the PSICQUIC View.
What is PSICQUIC?
PSICQUIC is an effort from the HUPO Proteomics Standard Initiative (HUPO-PSI) to standardise the access to molecular interaction databases programmatically.
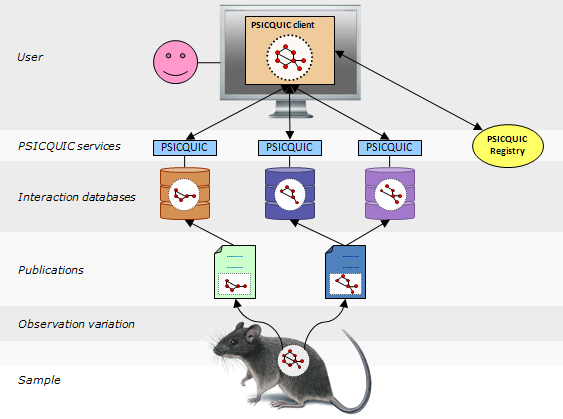
Basically, PSICQUIC specifies:
- A standard web service with a well-defined list of methods, accessible using SOAP or REST.
- A common query language (MIQL)
Many interaction databases are already implementing PSICQUIC. You can check the Registry to see the most up to date list of providers and their status.
If you are a user in need of a convenient and standard way to fetch molecular interaction data from different sources, check the following section Information for Users. If you are a service provider willing to facilitate your data so it can reach more users with very little effort, check the section Information for Molecular Interaction providers below.
Information for Users
PSICQUIC is a web service so it is meant to be use in machine-to-machine communications. However, you can get PSI-MITAB data directly in your browser by using the REST access, like in this example.
- How to retrieve molecular interactions: using SOAP or REST
- List of PSICQUIC Service Providers.
- There are two available JAVA clients for PSICQUIC. If you are a JAVA programmer start using PSICQUIC right away, find the documentations about the clients here.
- A Bioconductor package has also been developed for PSICQUIC. You can find more documentation here.
- You can find more examples, including examples in Perl or Python.
- The MIQL (Molecular Interaction Query Language) reference that can be used to perform more complex queries.
- If you want to see a real use case of PSICQUIC, check the PSICQUIC View application (then just click on Search). It uses PSICQUIC behind the scenes.
- You can also find the different PSICQUIC services in BioCatalogue.
- You can use PSICQUIC directly from the latest Cytoscape version. Check this page if you want to know how.
- Other projects are using PSICQUIC too.
Information for Molecular Interaction providers
Implementing PSICQUIC is relatively simple. If you can generate a PSI-MITAB file (versions 2.5, 2.6, 2.7 or 2.8), you can start using the Reference Implementation of PSICQUIC right away. For instructions, check these links:
- How to install the PSICQUIC Reference Implementation: step by step guide for molecular interaction providers.
Citing PSICQUIC
To cite PSICQUIC please use:
Aranda, B. et al. PSICQUIC and PSISCORE: accessing and scoring molecular interactions. Nat Meth 8, 528-529 (2011).
doi:10.1038/nmeth.1637 http://www.nature.com/nmeth/journal/v8/n7/full/nmeth.1637.html